Cutting-Edge Research: Exploring Bacterial-Phage Interactions with Microfluidics and Deep Learning
In the face of a global rise in antimicrobial resistance, we are at a critical juncture in the search for alternatives to traditional antibiotic treatments. Bacteriophages, viruses harmless to humans but deadly to bacteria, offer a promising avenue for potential treatments, a concept known as phage therapy. Recent advances in single-cell microscopy techniques have opened new doors for researchers, enabling the development of innovative methods to evaluate the dynamic interplay between bacteria and phages.
Here, we present two paper two papers that are recently published on Frontiers Microbiology and Frontier in Lab on Chip Technologies:
Our first approach (1) combines droplet microfluidics with time-lapse fluorescence microscopy to meticulously analyze the growth and lysis dynamics of Escherichia coli bacteria when exposed to phages. We investigated three distinct phage species: T7, MS2, and Qβ. By employing microfluidic trapping devices that generate and immobilize picoliter-sized droplets, we established a controlled environment for observing bacterial growth and lysis. This setup allowed us to track the temporal changes in bacterial populations for up to 25 hours, unveiling valuable insights into bacterial growth rates and the speed of phage infection.
In the long run, our research endeavors aim to propel the field forward by enhancing our understanding of the genetic and molecular mechanisms underpinning rapid phage-induced lysis. This knowledge will be invaluable in preventing bacterial resistance to phages and in determining the critical factors that influence the success of phage therapy.
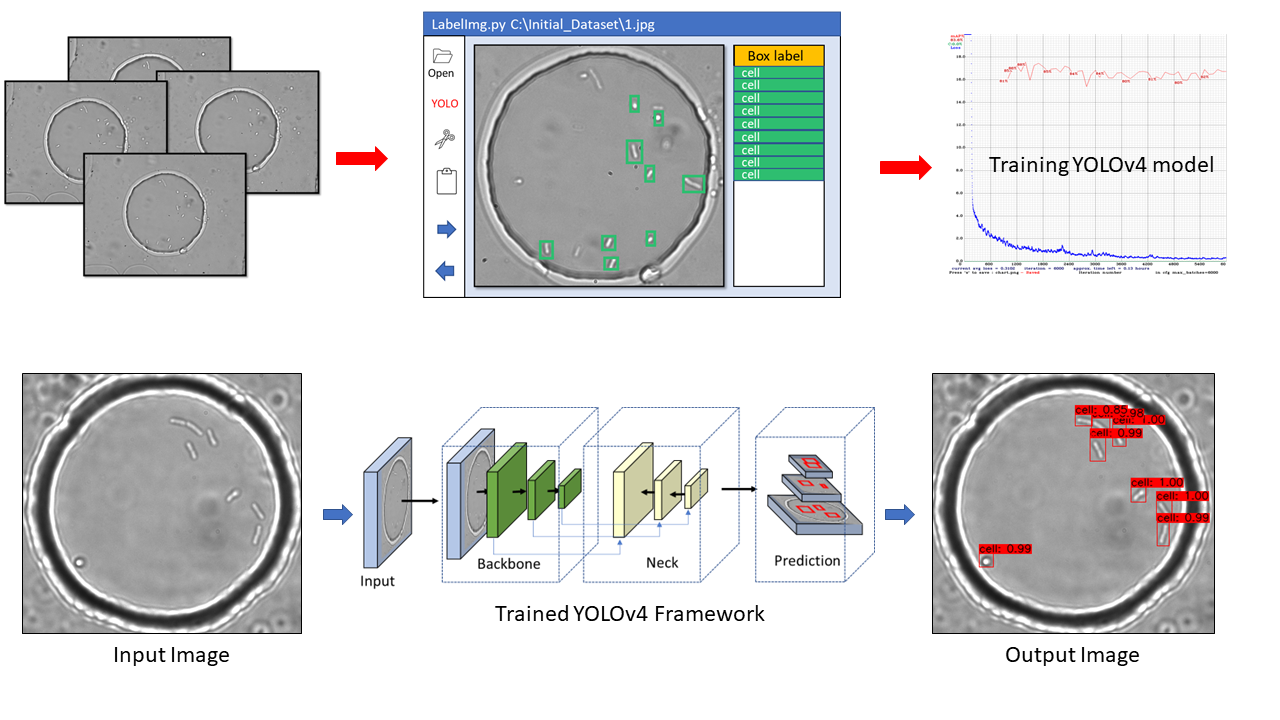
Label-free analysis of bacterial growth and lysis at the single-cell level using droplet microfluidics and object detection-oriented deep learning Link
Accurate identification and counting of small populations of motile bacterial cells hold significant implications for various fields, including food safety, infectious disease diagnostics, and antimicrobial research. However, the challenge remains in developing straightforward, rapid, and precise methods for counting single bacterial cells. We present a label-free solution (2) for counting and localizing freely swimming bacterial cells within microfluidic picolitre droplets.
Our innovative approach leverages the object detection-oriented YOLOv4 deep learning framework to identify bacterial cells from bright-field images captured using an automated Z-stack setup. With an impressive average precision of around 84%, the neural network is capable of recognizing the distinctive morphology of Escherichia coli cells. This breakthrough enables us to accurately pinpoint individual cell division events, providing an unprecedented opportunity to study stochastic bacterial growth, even from initial populations as low as a single cell.
Our research doesn't stop there. We also investigate single-cell lysis in the presence of T7 lytic bacterial viruses, commonly known as phages. The remarkable precision in cell counting facilitates the visualization of bacteria-phage interactions over hours, opening doors to comprehending phage life cycles within confined environments.
These studies mark significant progress in advancing our knowledge of bacterial- phage interactions, offering a promising avenue for future research and applications in various scientific and industrial fields.
Investigating bacteria-phage interaction dynamics using droplet-based technology Link